Introduction
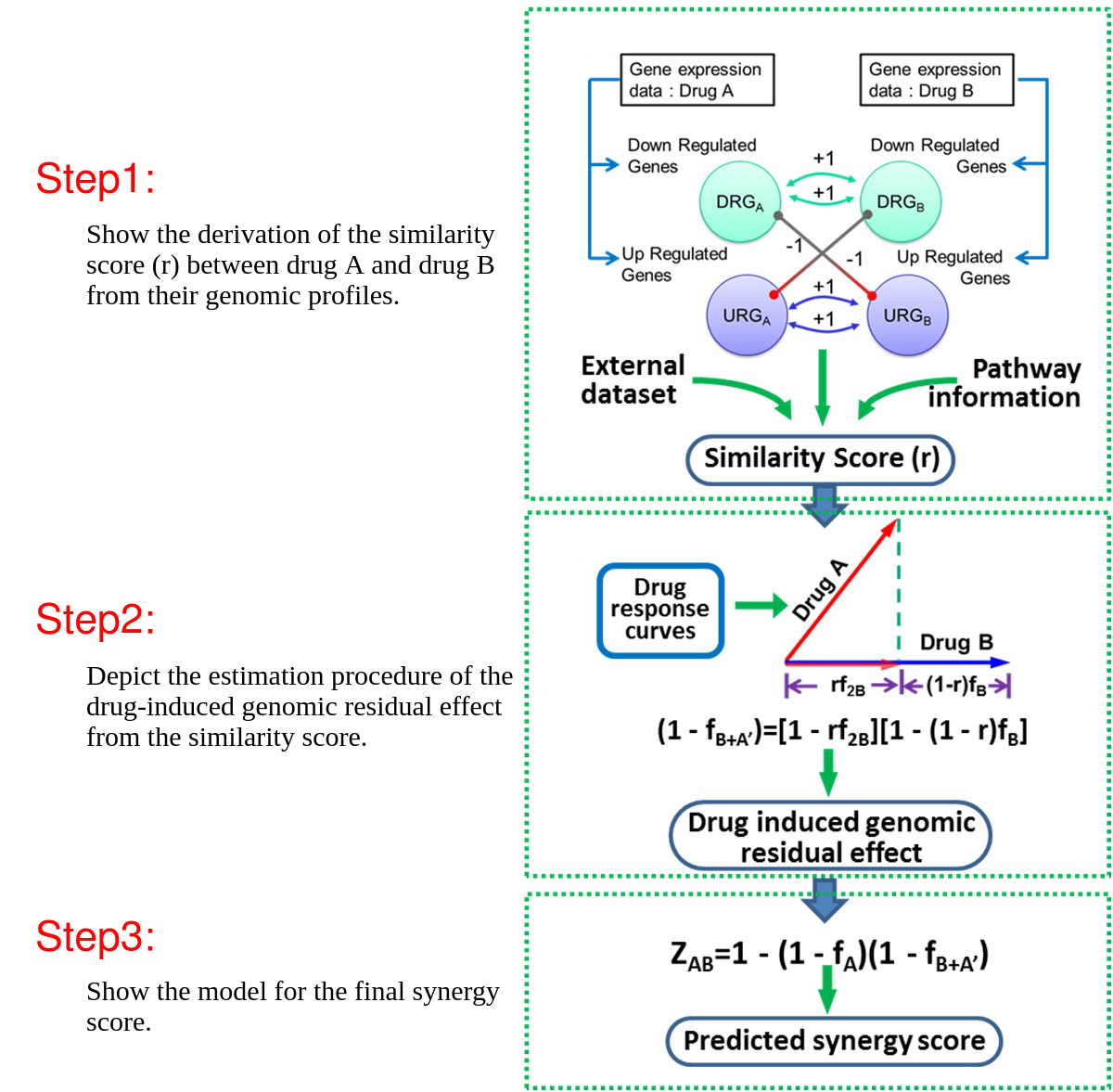
The Drug-Induced Genomic REsponse Model (DIGREM) was developed to predict compound pair synergistic effect. It uses drug-treated gene expression data as input, and outputs the predicted scores of synergistic effects and rankings of drug combinations.
The promise of multi-drug therapies for cancer has evoked renewed interest in high-throughput methods to identify effective drug combinations. Although a cell-based, large-scale single-drug screening strategy is well established, its extension to drug combination screening has become cost-prohibitive due to the sheer number of dugs to be tested. Our group aims to computationally predict the therapeutic effects of drug combinations, and thereby enable in silico screening of large repertories of drug combinations to prioritize potential effective multi-drug therapies for further experimental validation. This could dramatically revolutionize the current procedure of multi-drug therapy discovery and lead to the rapid identification of new combination therapies.
DIGREM integrates three methods (DIGRE, IUPUI_CCBB and gene set-based methods) to predict drug combination effects by explicitly modeling the drug response dynamics and gene expression changes after individual drug treatments.
The DIGRE model developed by our group won Best Performance in the National Cancer Institute’s DREAM 7 Drug Combination Synergy Prediction Challenge, an international crowdsourcing-based computational challenge for predicting drug combination effects using transcriptome data. This challenge’s blind-assessment of submitted computational models revealed that the prediction of drug pair activity from DIGRE was significantly consistent with the vast majority of the organizers’ experimental validations. In addition, we further validated our DIGRE model using another experimental dataset.
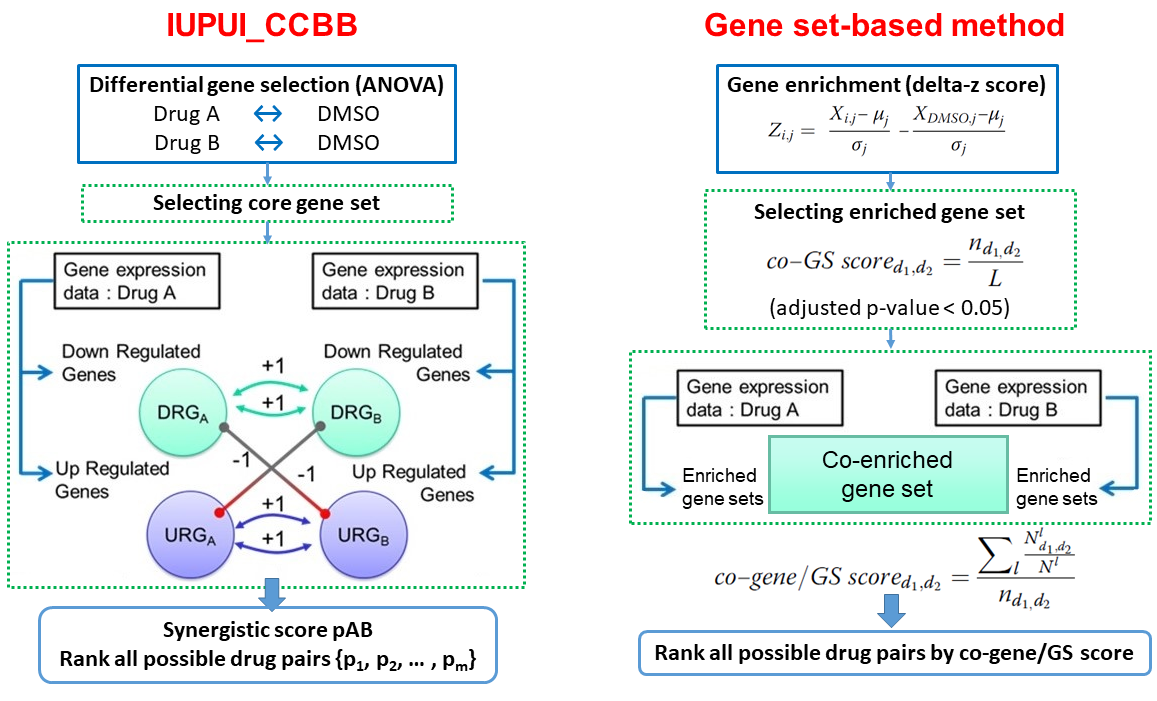
IUPUI_CCBB is the second-ranked method from the DREAM 7 challenge. It has a workflow similar to DIGRE's but does not consider gene-gene interaction or dose response curve. A statistical test is applied to identify commonly disturbed genes in a core gene set by two drugs, therefore replicates are required to ensure statistical power.
The gene set-based method is another recently developed method aiming the same question. Rather than looking at individual genes or a defined gene set, it calculates a gene set enrichment score across all gene ontology and oncogenic signature gene sets, and ranks drug pairs by an average percentage of commonly disturbed genes with the co-enriched gene sets.
DIGREM could potentially be used for the large-scale discovery of effective drug combinations for further experimental validation, possibly leading to the rapid identification of new therapies for complex diseases.
DIGRE model workflow
IUPUI_CCBB and gene set-based method workflow
Citing the tool:
Zhang, M., Lee, S., Yao, B., Xiao, G., Xu, L., & Xie, Y. (2018). DIGREM: an integrated web-based platform for detecting effective multi-drug combinations. Bioinformatics.Link
