Start analysis
To start using GeNeCK, prepare your gene expression data, and select your preferred
methods to construct the gene network.
>>> Demo
1. Select an algorithm |
|
Network Inference Method
GeNeCK provides 8 network inference methods that use distinct statistical
strategies:
1) partial correlation methods (GeneNet, NS, SPACE);
2) likelihood methods (GLASSO, GLASSO-SF);
3) information theory-based methods (PCACMI, CMI2NI); and
4) Bayesian methods (BayesianGLASSO).
Incorporate Hub Gene
Gene networks usually have scale-free characteristics. In other words, there are usually
a few hub genes regulating many others. EGLASSO and ESPACE
are two methods that incorporate the prior knowledge of hub genes to enhance network
inference. In these extended methods, during the covariance estimation, an additional
tuning parameter is introduced to reduce the penalty on edges connected to hub genes.
Integrative Method
Ensemble-based network aggregation (ENA) is an approach to combine network
reconstructed from different methods, which borrows the strength from different strategies
and generally yields good results.
2. Upload data & set parameters |
|
Gene expression data
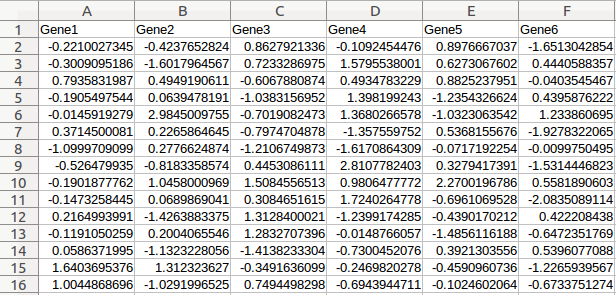
Gene expression data (e.g. microarray and RNA-Seq) monitors the transcription activities of different genes in a cell simultaneously, which is the foundation of all statistical algorithms. >>> Example
Method parameters
GeNeCK offers the flexibility for user to specify the value of parameters for
different methods. These parameters usually control the sparsity of the constructed network.
Though they may share the same name ($\lambda$ or $\alpha$), they have different and
sometimes opposite meanings in the context of different methods. Refer to each method to
choose an appropriate value.
Hub genes
Incorporating prior hub gene information can enhance the network inference, but make sure
those hub genes has real biological support. User can change the corresponding parameters in
EGLASSO and ESPACE to control the confidence level for hub genes.
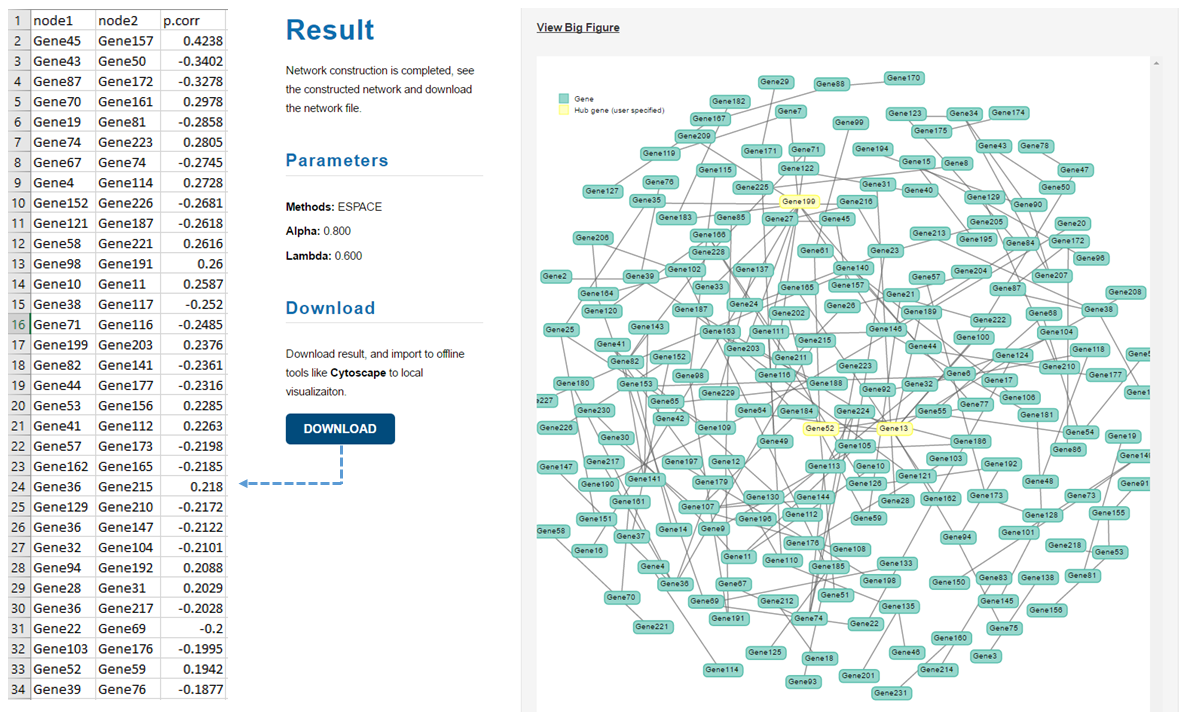
3. Visualize & download |
|
After you submit your job, you will be directed to a waiting page. You are free to leave and come back to check the job status at any time. If you provide your e-mail address, the link will also be sent to you. Once the job is done, the constructed network will be displayed on the page. You can also download the result that contains all the estimated edges. Each row represent two connected nodes. >>> Example