Upload File Requirements
- Your file MUST be comma-separated csv file
- Your file extension MUST be .csv
- Your file size MUST be smaller than 2.5MB
The CSV file format is below.
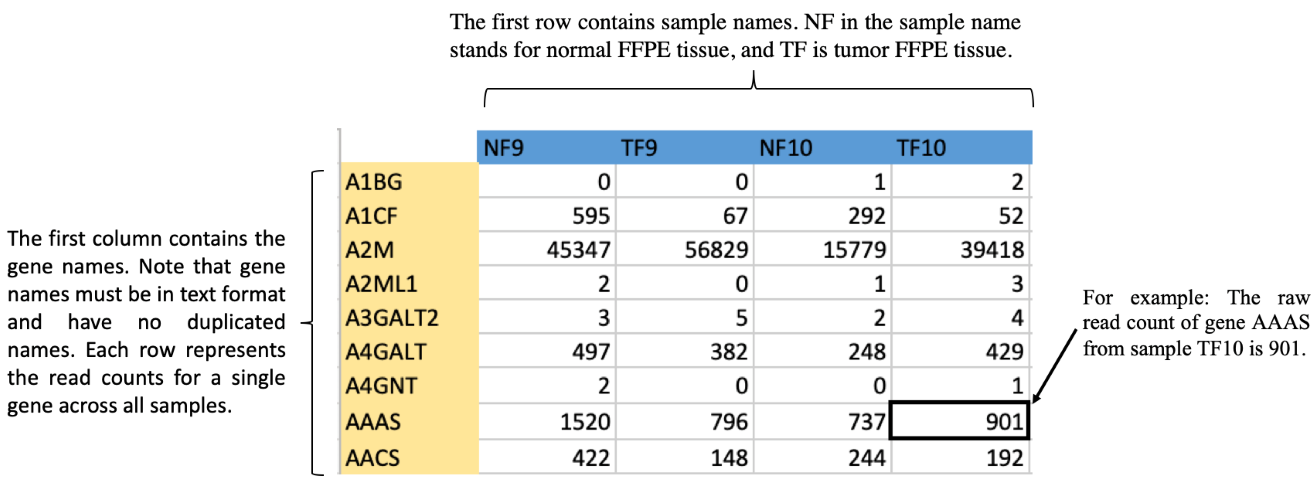
Download Example File
DESeq normalizes the raw count data by using (estimated) scaling factors and is implemented in R package DESeq. First calculate the ratio of the read count over the geometric mean across all samples for each gene. Then estimate the scaling factor by the median ratio within each sample.