Upload File Requirements
- Your file MUST be comma-separated csv file
- Your file extension MUST be .csv
- Your file size MUST be smaller than 2.5MB
The CSV file format is below.
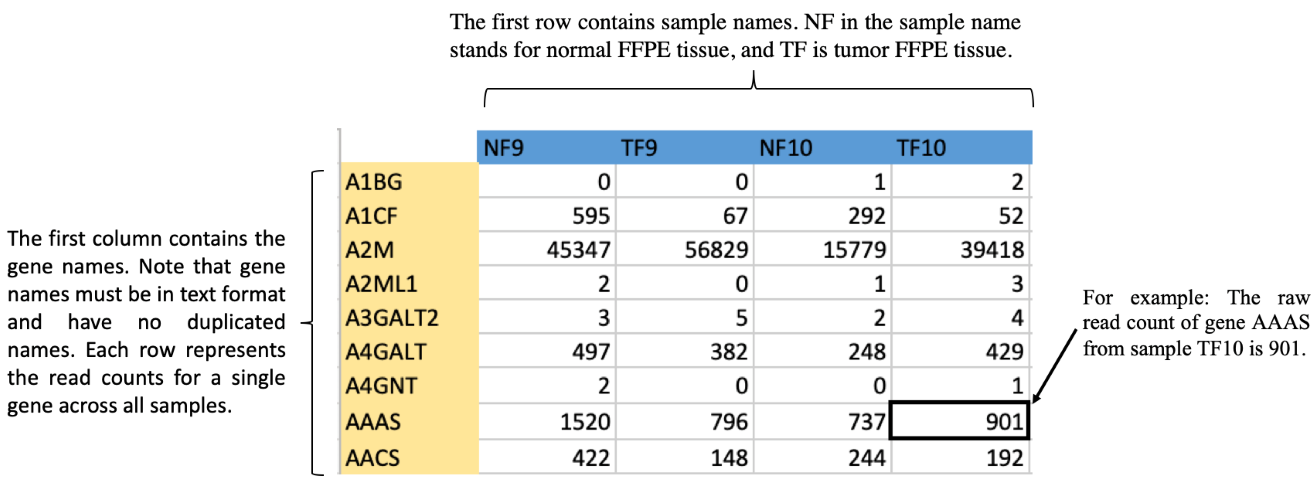
Download Example File
SMIXnorm is a simplified normalization method based on MIXnorm. The main simplification is to model the expressed genes by normal distributions without truncation. It is generally reasonable to assume that the densities for negative values are negligible such that the truncation makes no effect on modeling the expressed genes. SMIXnorm achieves comparable performance as MIXnorm, while greatly reduces the computation costs.
SMIXnorm takes the raw count data as input and output the normalized expression matrix in natural logarithm scale. The default maximum number of iterations is 15 and the stopping criteria is 0.01. The exact SMIXnorm uses the conditional probabilities of genes being expressed or not to produce the normalized data. The approximate SMIXnorm uses a cut-off value of 0.5 on those probabilities to classify genes as expressed or not. Non-expressed genes will be normalized to 0 by approximate SMIXnorm.